Molecular and Cellular Modeling
The dynamic behavior of biological systems arises from complex interactions between molecular components within cells. Our group applies mathematical modeling to simulate and predict molecular and cellular processes that are critical in understanding how these systems operate under normal and disease conditions. We focus on developing Ordinary Differential Equation (ODE) models, agent-based models, and stochastic simulations to investigate key biological phenomena such as viral replication, cellular signaling, and the response to pharmacological interventions. These models help to elucidate how molecular perturbations can affect cell behavior, providing insight into novel therapeutic targets for diseases like cancer, viral infections, and age-associated degenerative disorders. Through sensitivity analysis and model optimization, we aim to predict system responses and uncover new intervention points to modulate biological pathways in disease contexts.
Virus-Host Interaction Models

Viruses rely on host cellular machinery for their replication and survival. Understanding these interactions is key to developing novel antiviral strategies, especially for viruses with high mutation rates. Our group develops dynamic models to quantify and predict how viruses hijack host cell processes and how these interactions evolve over time.
By integrating data from nanopore sequencing, mass spectrometry, and time-resolved viral replication measurements, we simulate the virus-host interface to identify potential host factors that can be targeted to inhibit viral replication. One of our current focuses is on building models of virus-host interactions for the SARS-CoV-2 replication cycle, identifying non-structural proteins (NSPs) that could be viable drug targets. These computational models provide a systems-level understanding of viral infections and facilitate the discovery of new therapeutic interventions by exploiting host-pathogen dependencies.
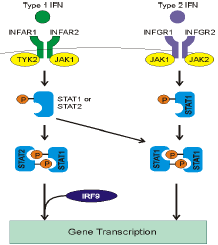
Immune System and Cellular Response Modeling
The immune system’s dynamic response to infections involves complex signaling pathways and feedback mechanisms that determine the outcome of host-pathogen interactions. We focus on modeling the intricate immune responses triggered by viral infections, particularly how immune cells detect and respond to pathogens through molecular sensors like RIG-I and Toll-like receptors (TLRs). Our mathematical models of innate immunity, which incorporate pathways like NF-κB and interferon signaling, help to predict how the immune system manages the balance between viral replication and immune activation. By simulating the interplay between viral replication and the cellular immune response, we can pinpoint how immune evasion strategies by viruses, or failures in immune regulation, contribute to disease progression. This work is crucial for designing more effective antiviral therapies and vaccines that enhance the body’s natural defenses against infection.
Systems Biology of Cancer
Cancer is characterized by extensive molecular reprogramming that affects multiple biological networks. Our research in
systems biology of cancer aims to understand how dysregulated signaling and gene networks contribute to tumor development, progression, and resistance to treatment. We develop computational models of key oncogenic pathways, such as those involving EGF receptor trafficking, and examine how perturbations in these pathways drive uncontrolled cell growth and evade apoptosis. Recent projects focus on integrating multi-omics data—genomics, proteomics, and transcriptomics—to uncover the underlying molecular mechanisms that enable cancer cells to thrive. These models are essential for predicting the effects of potential therapeutic interventions and for identifying drug targets that can disrupt the systemic networks that fuel cancer growth and metastasis.
Ageing and Age associated Diseases

Aging leads to systemic dysregulation of cellular functions, contributing to the onset of multiple age-related diseases, including Alzheimer’s disease, cardiovascular disease, and cancer. In our group, we use high-throughput data from genomics, transcriptomics, and proteomics to build models that explore the molecular underpinnings of aging.
Our research combines network biology and multi-scale modeling to study processes such as telomere shortening, genomic instability, and protein misfolding, all of which contribute to cellular senescence and age-associated dysfunction. We use network inference techniques and pathway analysis to map out the regulatory networks involved in aging and to identify key nodes that could be targeted to delay or reverse aspects of aging. In particular, our modeling of age-related diseases is focused on understanding the complex interaction between genetic predispositions and environmental factors that accelerate aging. These insights provide a foundation for developing novel interventions aimed at extending healthspan and combating age-associated diseases.
